The general procedure for FISH involves fixing samples of chromosomes or human tissue onto a piece of glass known as a slide (it slides into place on the viewing platform of a microscope when the sample is ready to examine). To prepare the tissue on the slide for hybridization, it is treated with chemicals to permeabilize (open up) the cells and denature the DNA, so that the probe can more readily hybridize.
A complementary nucleic acid probe is prepared and labeled, either by radioactivity or by fluorescence. A hybridization solution containing the probe is sloshed over the slide so that the probe can hybridize with the sequence of interest. Factors such as the temperature, pH, and salt concentration can be changed to control the specificity of the hybridization.
The excess probe is washed away. Then, stained using a contrasting color and the cells are viewed using several different techniques and the most common used is fluorescence microscope. See Figure 1 for a flow chart of FISH.
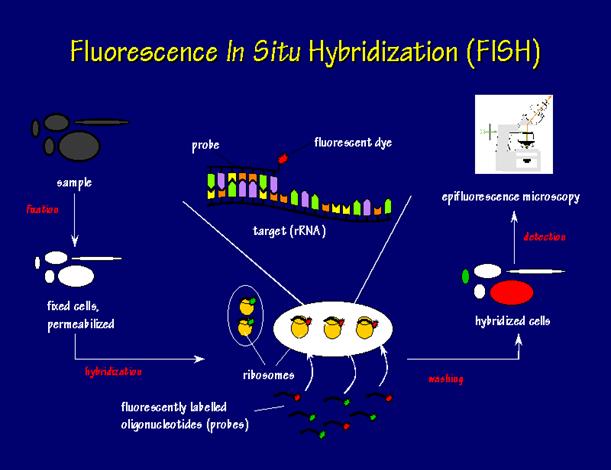
Figure 1: Flow chart of fluorescent in situ hybridization
First, a probe is constructed.
It should be labeled directly, where the reporter molecule is directly attached to the probe, or indirectly, where a specific antibody or labeled binding protein is used to detect another molecule that is attached to the probe sequence.
Fluorescin and Rhodamine can be used for direct fluorescently labeled probes, and biotin and digoxigenin can be used for indirect fluorescence labeling.
This can be done in a variety of ways, for example nick translation and PCR using tagged nucleotides.
Different kinds of probes:
· Oligonucleotide
· Cosmid
· Alpha satellite
· Painting
DNA FISH, probes include double-stranded DNA, single-stranded DNA, and synthetic oligodeoxyribonucleotides
For RNA FISH, the probe can be single-stranded complementary RNA, called a riboprobe, which is synthesized by reverse cloning (Polak et al., 1990)
Fixation and Denaturation of Material
The DNA or RNA must be fixed onto a glass slide for stability.

Next step is to break apart (denature) the double strands of DNA in both the probe DNA and the chromosome DNA.
This is done by heating the DNA in solution of formamide at a high temperature (70 C).
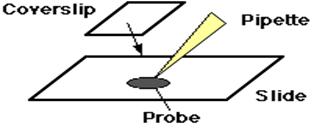
And, the probe is placed on the slide and a glass coverslip is placed on top.
Hybridization
The slide is then placed in a 37 C incubator overnight for the probe to hybridize with the target chromosome. The degree of specificity to which the probe hybridizes to the target sequence can be controlled by the design of the probe and the conditions of the buffer solution, including temperature, pH, and salt concentration. “High stringency” conditions will only allow hybridization of probes with very similar homology to the target sequence, while “low stringency” conditions will allow a probe to bind with less specificity.
Overnight, the probe DNA seeks out it is target sequence on the specific chromosome and binds to it. The strands slowly rejoin (reanneal).


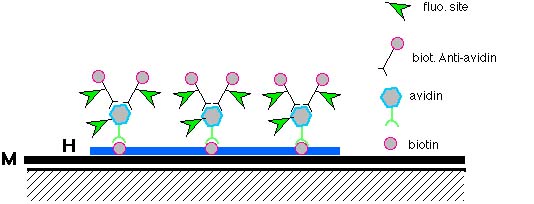
Visualization
The next day, the coverslip is removed, and the slide is washed in a salt/detergent solution to remove any excess of the probes. A differently colored fluorescent dye is added to the slide to stain all of the chromosomes so that they may then be viewed. Several different techniques can be used to view where the probe has hybridized with the sequence of interest. Light field microscopy is most common and can be used for radioactively labeled probes or probes labeled with peroxidase or alkaline phosphatase. Fluorescent microscopes are used to view fluorescently labeled probes. Digital imaging systems are also used and can process the images and do quantitative measurements
Actually, two (or more) different probes labeled with different fluorescent tags can be mixed and used at the same time. The chromosomes are then stained with a third color for contrast. This gives a metaphase or interphase cell with three colors which can be used to detect two different chromosomes at the same time, or to provide a "control probe" in case one of the other target sequences are deleted and a probe cannot bind to the chromosome.
Related Sites:
http://www.slh.wisc.edu/cytogenetics/procedures/fish/fish-method.php
http://www.accessexcellence.org/RC/VL/GG/fish.html

